tucca-rna-seq: An RNA-Seq Workflow
The tucca-rna-seq workflow is a comprehensive solution for RNA-Seq data
analysis. It provides a fast, automated pipeline for researchers and
bioinformaticians, regardless of their programming experience.
Originally developed for cellular agriculture research, this workflow is designed to be a robust, user-friendly toolset that streamlines RNA-Seq data analysis, ensuring accuracy, reproducibility, and scalability for any research field.
Our goal is to accelerate research by providing a powerful, accessible, and well-documented workflow. For those interested in the cellular agriculture origins of this project, you can learn more here.
Workflow Overview
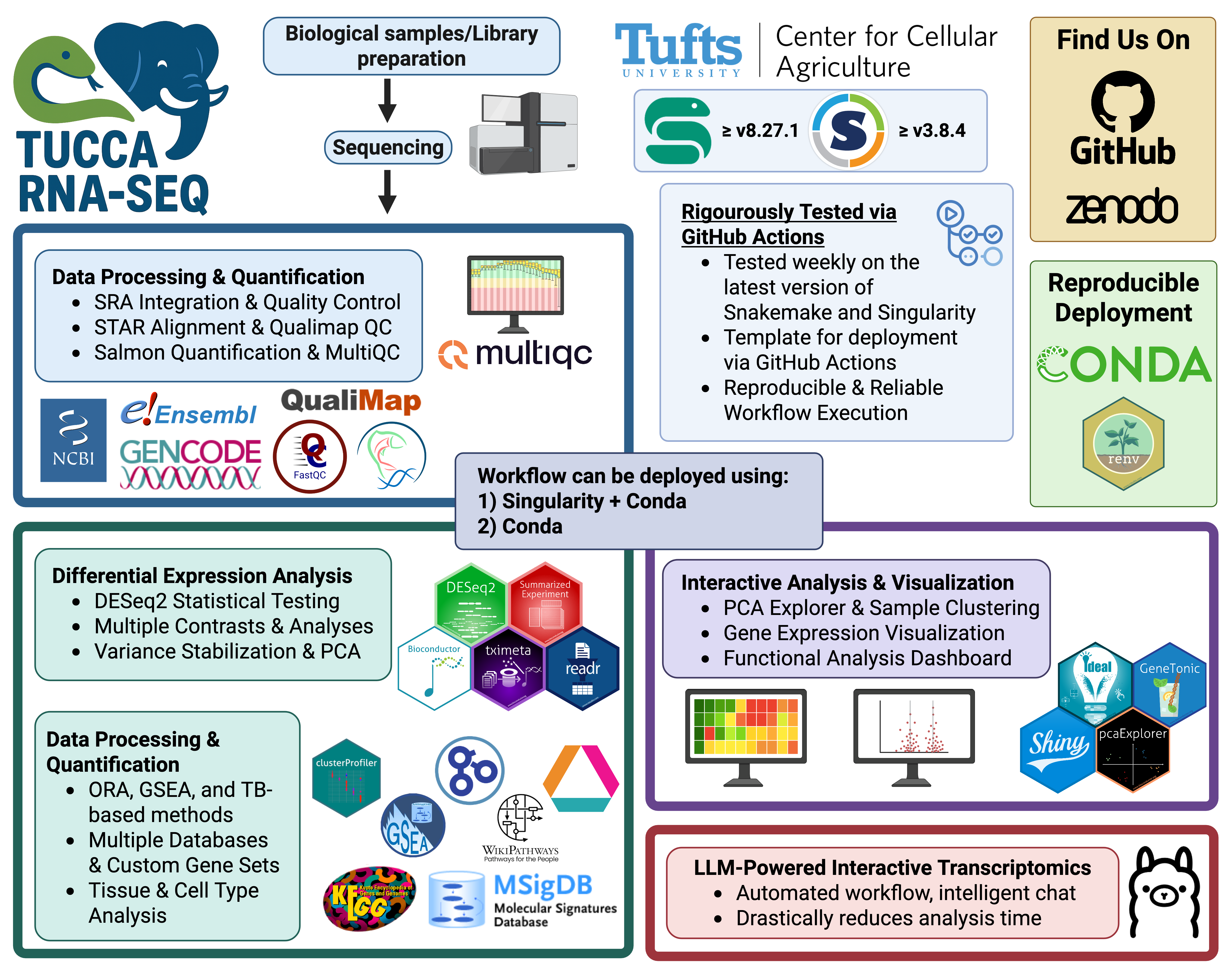
High-level overview of the tucca-rna-seq workflow.
Created in https://BioRender.com
Real-World Impact: A Case Study in Cellular Agriculture
This workflow has been instrumental in generating novel scientific insights for
published research. An early development version of tucca-rna-seq was pivotal
in a study on cellular adaptation for cultivated meat, enabling the discovery of
key biological mechanisms that govern how cells transition from adherent to
suspension growth.
This case study demonstrates how the workflow can be leveraged to translate raw sequencing data into a deeper biological understanding.
About the Workflow
The tucca-rna-seq workflow provides an efficient and seamless pipeline for
RNA-Seq data analysis. Key features include:
Comprehensive Data Processing
- Quality Control: Implements
FastQCandQualimapfor quality assessment with configurable parameters. - Quantification: Employs
Salmonfor fast, accurate transcript quantification, accounting for experimental attributes and biases. - Meta-Analysis: Aggregates results using
MultiQCfor a unified overview of data quality and processing metrics.
Advanced Differential Expression Analysis
- Statistical Tools: Leverages
DESeq2for robust differential expression analysis. - Flexible Configurations: Supports multiple experimental designs and contrasts in a single run, with configurable statistical parameters.
- Data Transformation: Provides variance-stabilizing transformations (rlog, vst) for downstream analysis.
Comprehensive Functional Enrichment
The workflow integrates multiple enrichment analysis approaches:
- Standard Analysis: GO, KEGG, and WikiPathways using
clusterProfiler. - MSigDB Integration: Access to 8 major gene set collections with custom GMT file support.
- SPIA Analysis: Topology-based pathway impact analysis.
- Harmonizome Database: Tissue-specific gene sets from various expression datasets.
- Custom Gene Sets: Support for user-defined gene sets and pathways.
Post-Analysis Interactive Tools
- RMarkdown Playgrounds: Interactive notebooks for data exploration and visualization.
- PCA Explorer: Interactive principal component analysis and sample clustering.
- GeneTonic Integration: Comprehensive enrichment analysis visualization with exportable figures.
- Custom Analysis: A framework for extending analysis beyond workflow outputs.
The integrated Shiny applications (GeneTonic, pcaExplorer) provide interactive visualization experiences that allow you to download plots and figures in multiple formats (PNG, PDF, SVG) directly from the apps.
High Reproducibility
- Environment Management: Employs
condaandrenvto replicate computational environments, ensuring consistency. - Container Integration: Uses Singularity/Apptainer containers for consistent execution environments.
- Schema Validation: JSON schema validation for configuration files.
- Version Control: Maintains version-controlled workflows with
Snakemakeandgitfor tracking and replication.
Scalability and Flexibility
- HPC Integration: Optimized for high-performance computing clusters.
- Multiple Execution Environments: Support for Slurm, LSF, Kubernetes, and cloud platforms.
- Modular Design: Easily extensible and customizable.
- Testing Infrastructure: Comprehensive CI/CD testing with GitHub Actions.
Modern Snakemake Integration
- Snakemake v8+: Built for the latest Snakemake with modern executor plugins.
- Executor Plugins: Dedicated plugins for different computing environments.
- Simplified Profiles: Easy-to-configure execution profiles.
- Resource Management: Intelligent resource allocation and job scheduling.
Workflow Rule Graph
The diagram below illustrates the rule graph (DAG) for the tucca-rna-seq
Snakemake workflow. Each node represents a workflow rule, and edges indicate
dependencies between steps. This provides a high-level view of how data moves
through the pipeline from raw input to final results.
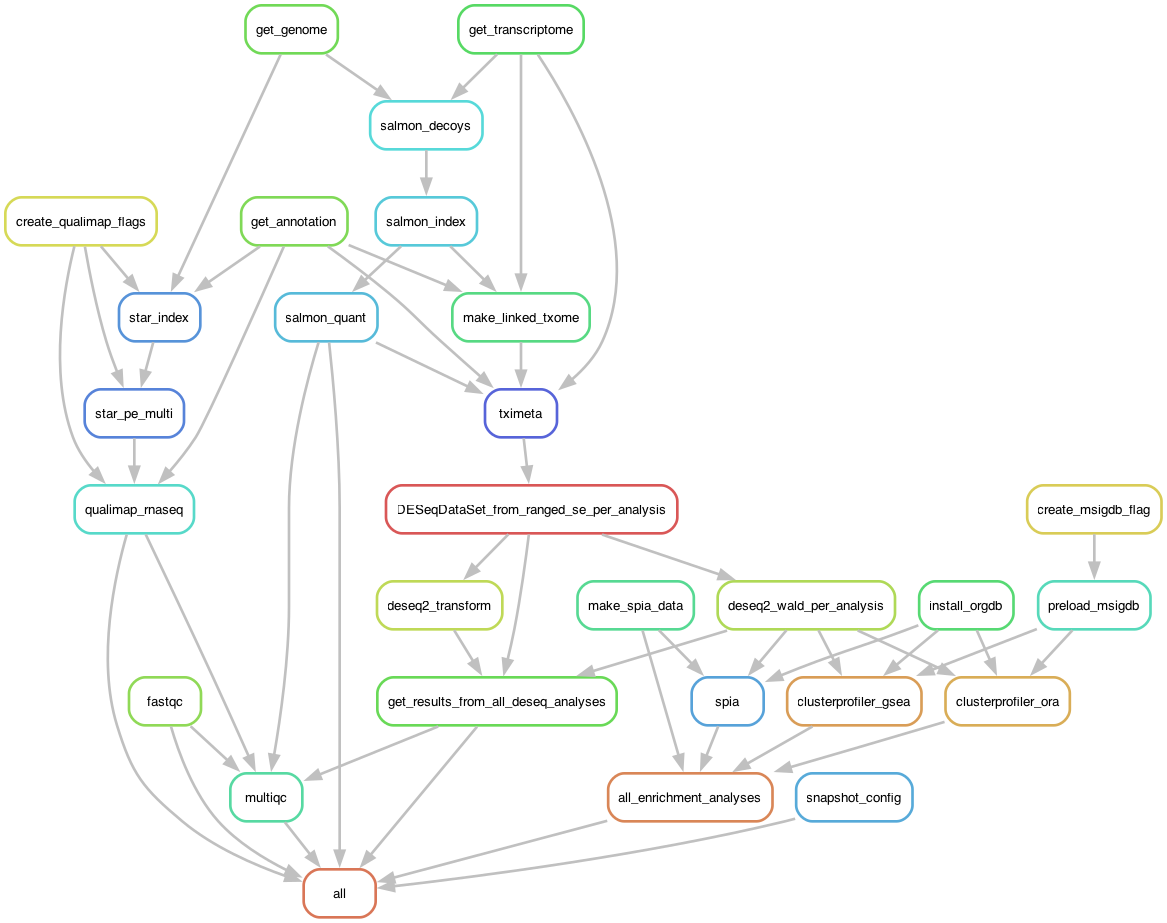
Rule graph (DAG) of the tucca-rna-seq Snakemake workflow.
Getting Started
- Data Collection: Why and how to collect the metadata you'll need for your analysis.
- Snakemake Primer: An introduction to the core concepts behind Snakemake and reproducible workflows.
- Deployment Strategies: An overview of the different platforms where you can run this workflow.
- Installation: How to install the required software for the workflow.
- Configuration: How to configure the workflow for your specific analysis.
- Running the Workflow: How to execute the workflow and monitor its progress.
Platform-Specific Guides
- Tufts HPC Quick Start: A guide for running the workflow on the Tufts HPC cluster.
- Tufts HPC Best Practices:
Advanced tips for using
tucca-rna-seqon the Tufts HPC. - VSCode Extensions: Recommended VSCode extensions for an improved user experience.
Additional Resources
- Reproducibility in Bioinformatics: Best practices for reproducible code and project management.
- R Extensions: Essential R packages and tools for data analysis.
Connect With Us
We're here to help! If you have questions, feedback, or need assistance, feel free to reach out through our social channels:
Alternatively, visit our GitHub Repository to explore more of TUCCA's projects.