Snakemake Primer
Scientific results need to be reproducible, but this can be challenging in computational biology where software versions and system configurations vary. Our workflow is built using a combination of tools that solve this problem.
Snakemake: A Cookbook for Bioinformatics Workflows

At its core, our workflow is managed by Snakemake. Think of a
Snakemake workflow (defined in a file called a Snakefile) as a
detailed recipe for a complex scientific analysis. Each step in the analysis,
like aligning reads or counting genes, is a "rule" in the recipe. Each rule
clearly defines its inputs (the ingredients) and the outputs it will
produce (a prepared component of the final dish).
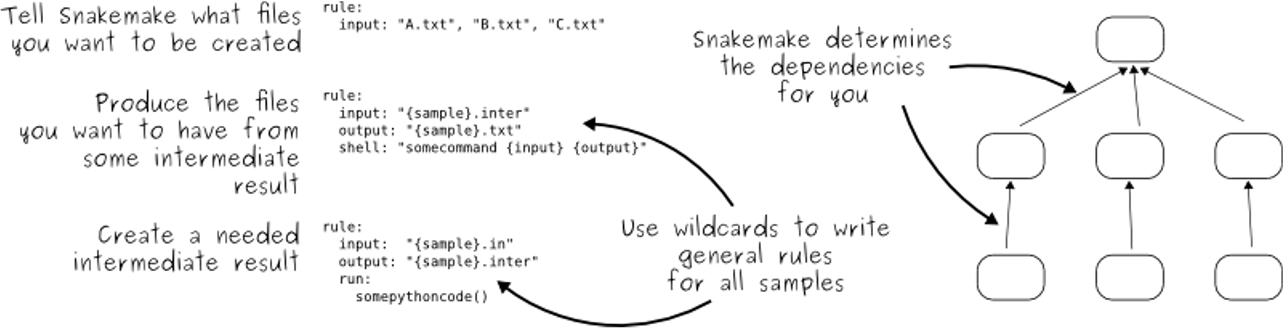
Snakemake is the master chef that reads this recipe. It understands the
dependencies between all the steps and uses the Snakefile to intelligently
automate the entire analysis, providing several key advantages:
- Automation: It automatically determines the correct order to run each rule. This prevents human error and saves you from manually executing dozens of commands.
- Efficiency: It can run multiple independent rules in parallel, dramatically speeding up the analysis, especially on multi-core computers or HPC clusters.
- Reproducibility: If an input file or a parameter is changed, Snakemake knows exactly which steps need to be re-run, ensuring the results are always up-to-date.
Before executing anything, Snakemake first examines the entire workflow, noting all the inputs and outputs of every rule. It uses this information to build a map of job dependencies called a Directed Acyclic Graph (DAG). This DAG is the master plan that determines exactly what steps need to be run and in what order.
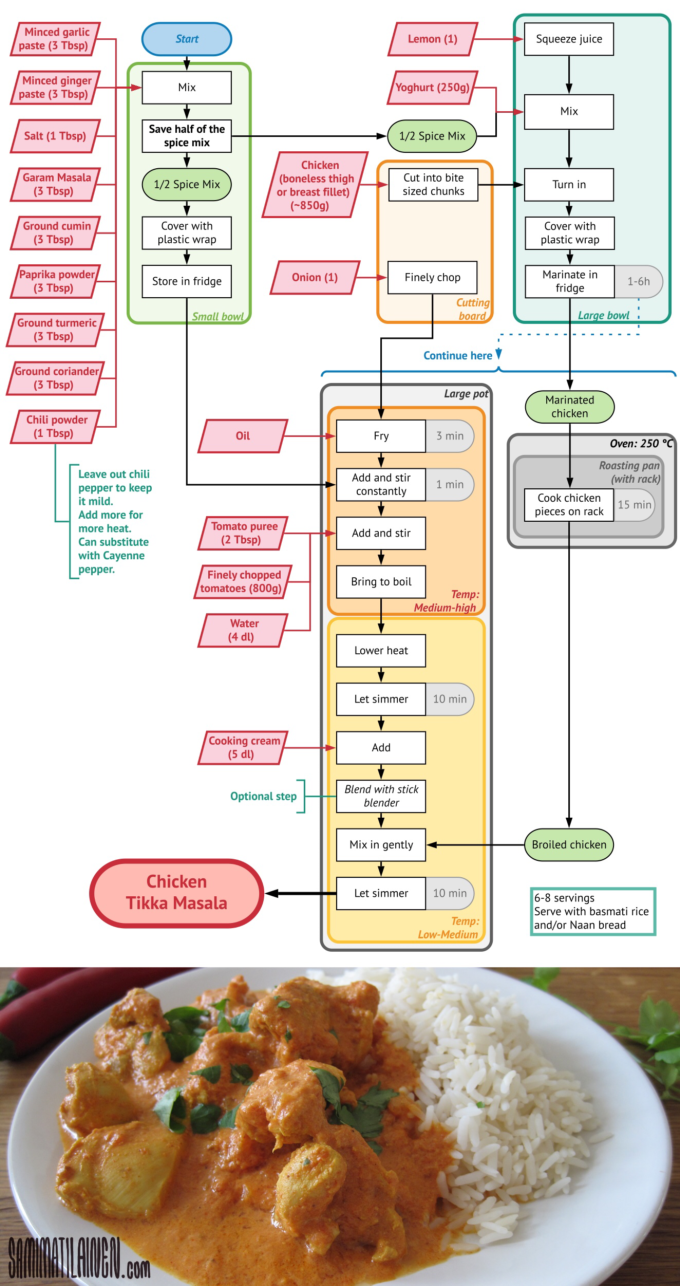
Chicken Tikka Masala as a DAG
A recipe has a list of ingredients (inputs) and steps that must be followed in order to produce a dish (output). This is an example of a directed acyclic graph (DAG) for a chicken tikka masala recipe.
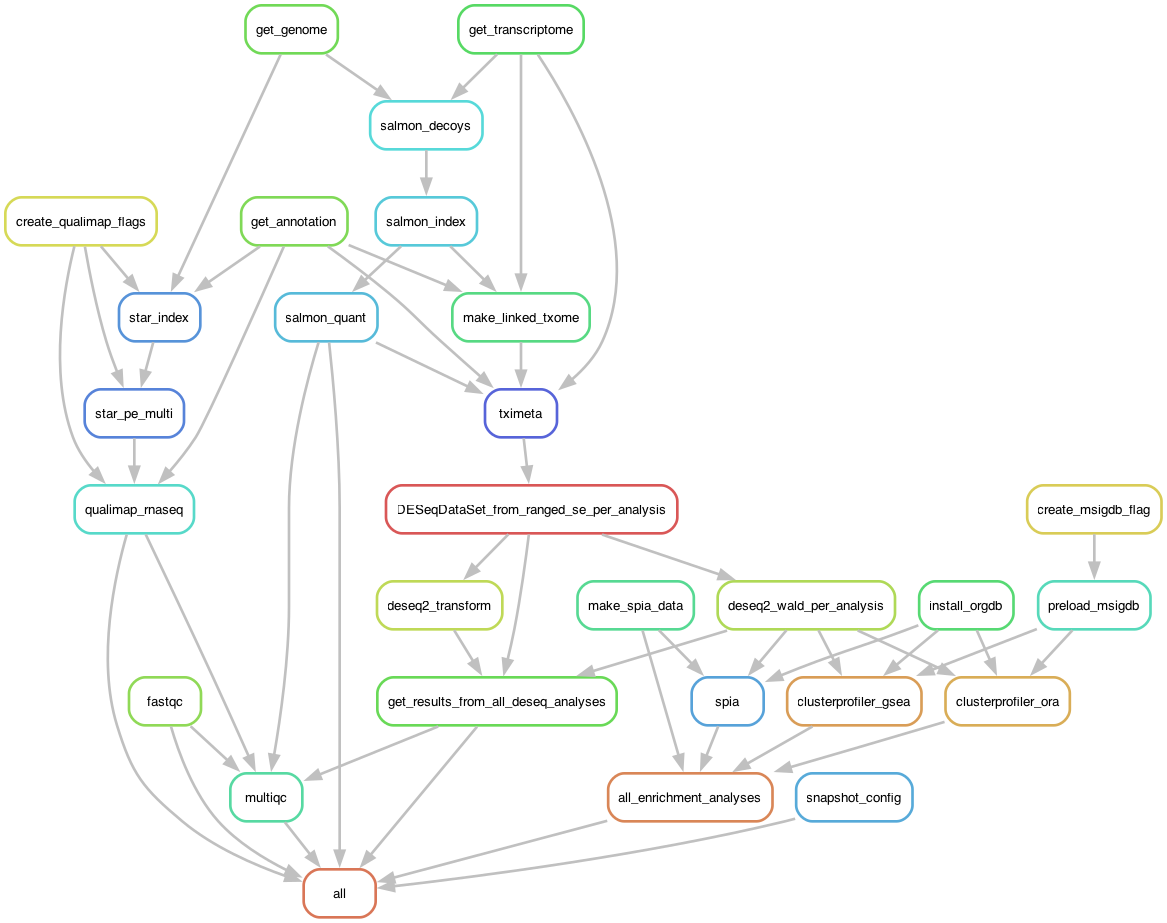
The tucca-rna-seq Workflow DAG
Snakemake defines each step as a "rule" with defined inputs and outputs, creating a directed acyclic graph (DAG) from your initial raw reads to the workflow's various outputs.
Snakemake Links Docs and API
The workflow management system that orchestrates the analysis, using Conda to create reproducible environments for each step.
This is the most important page to visit when learning how to execute, debug, and configure your workflow.
For more advanced needs, or when information isn't in the main docs, the API reference is useful.
Learn Snakemake (Recommended Tutorials)
The Reproducibility Problem: Why Your "Oven" Matters in Bioinformatics
Imagine two people trying to bake the exact same cake using the same recipe. One person uses a modern convection oven, while the other uses an older gas oven. Even with the same ingredients, the different ovens (their "environment") can produce slightly different cakes.
This is a common problem in bioinformatics. A script that works perfectly on one computer might fail or produce different results on another. This is because the "environment"—including the operating system and the specific versions of software tools—is different.
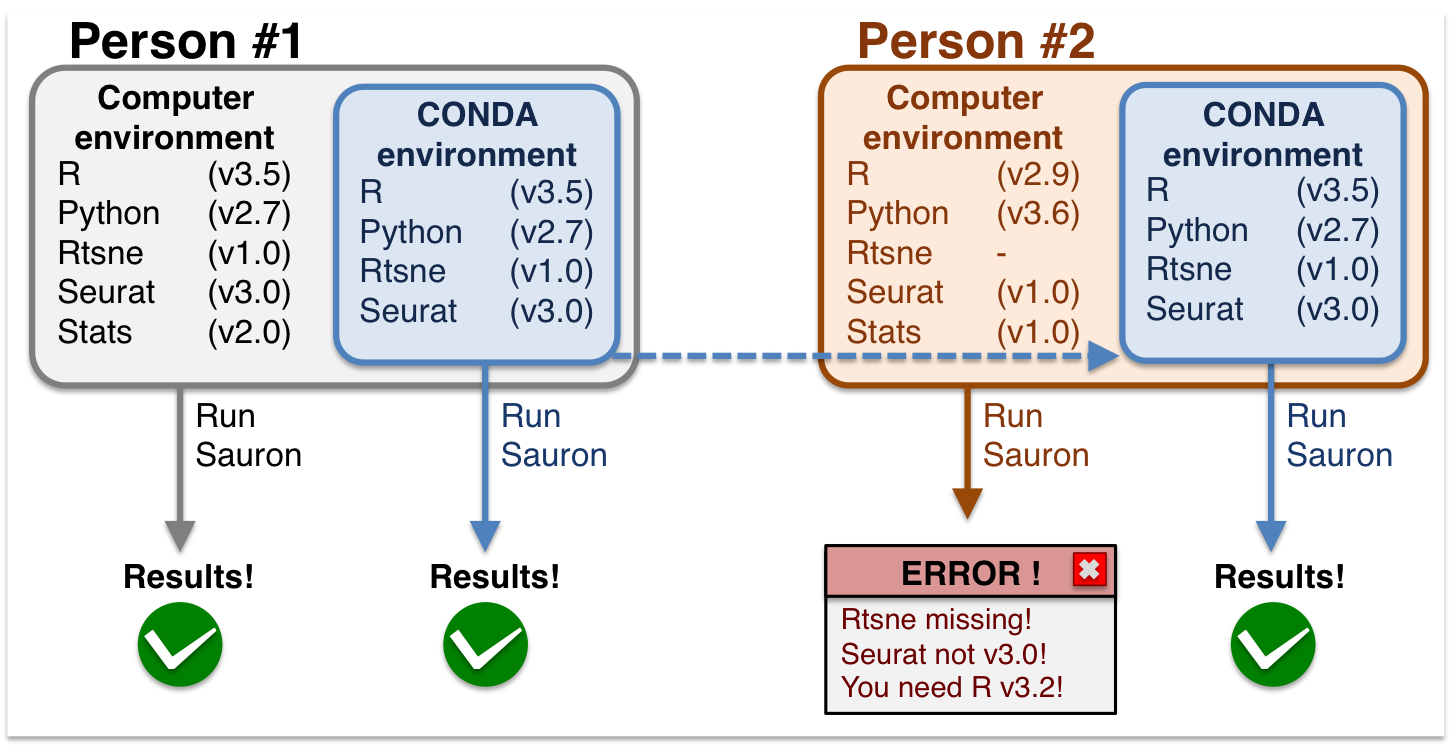
The "It Worked on My Machine" Problem
In this example, Person #1 can run the analysis successfully. When Person #2 tries to run the same analysis on their computer, it fails because of different software versions. This is a classic reproducibility issue.
Conda: The Universal Shopping List
To begin to solve this, we can use a package manager like Conda.
If the Snakefile is our recipe, think of Conda as a universal, automated
shopping list. For each step in our recipe, Conda reads a list of required
software "ingredients" (e.g., a specific version of a read aligner or a quality
control tool) from a dedicated environment.yaml file.
It then automatically:
- Finds the exact versions of the software specified.
- Installs them into an isolated, self-contained "environment".
- Ensures that the software for one step doesn't conflict with the software for another.
This way, when Person #1 shares their Snakefile and their Conda environment
files, Person #2 can recreate the exact same software setup, making the
analysis much more likely to run successfully.
However, Conda only manages the software inside a given operating system. What if the two computers have fundamentally different operating systems (e.g., Windows vs. macOS vs. Linux)? Tiny differences in system-level libraries can still cause problems. This brings us to the ultimate solution: containers.
Containers: The Perfect Kitchen
To solve the environment problem, we use containers.
A container is like a standardized kitchen. It doesn't just contain the ingredients (the code and data), but also the exact kitchen appliances (specific software versions) and the recipe book (the operating system) needed to make the meal perfectly every time.
When you run the workflow using a container, you're running it in a consistent, pre-configured environment. This ensures that the analysis will produce the exact same results, regardless of what machine it's run on. This is the gold standard for reproducible science.
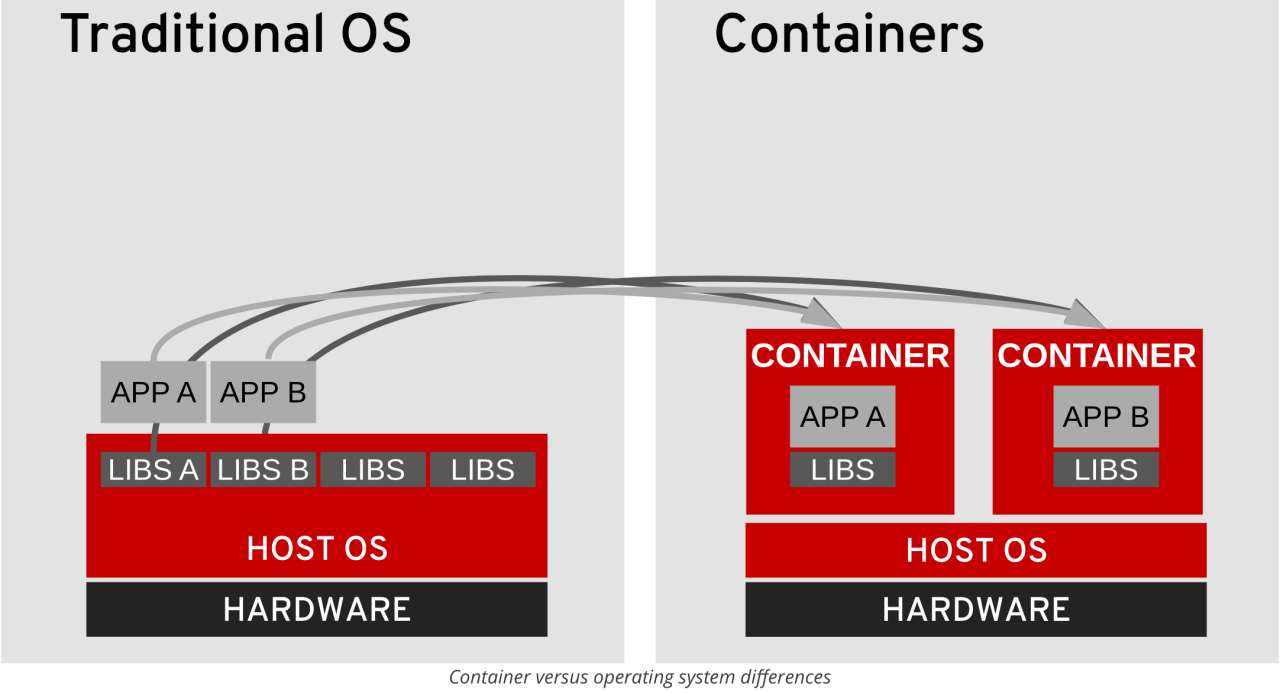
How Containers Create Consistency
On the left, traditional applications rely on the host operating system's libraries, which can vary. On the right, containers bundle their own libraries, ensuring the application always runs in the exact same environment.